
When I run the search engines in command line, all eight seem to work (returns usage instructions). I noticed that one error in Tide states "Load spectra:Error in MGF file: bad m/z or intensity value." Could the problem be in the MGF file? The same error resulted as with my custom database, so it seems unlikely that my database is the (only) problem, at least with OMSSA. I did a quick test to see if the database format could be the problem by repeating the identical search with the UniProt database using OMSSA. While running OMSSA through SearchGUI, I noticed that "omssacl.exe (32 bit)" was running in Task Manager.
#Peptideshaker download download#
They lead to the Microsoft "Top Download Categories" page. The links on the SearchGUI troubleshooting page under "OMSSA for Windows" don't seem to lead to the right place anymore. Sorry about the confusion! This leaves OMSSA, Comet and Tide that do not work on the PC, and MyriMatch, OMSSA, Comet and Tide on Linux (I haven't tested MS Amanda on Linux yet, will do so when I can).
#Peptideshaker download Pc#
MS Amanda is also working on PC now (I must have done something wrong last time, was also with an older version). I forgot to mention that Andromeda worked, but I decided not to use it for my analysis as discussed in a previous issue here. The searches that failed gave a variety of error messages. Have you used PathSettingsCLI to set the temporary paths to folders you have write access to? Path related issues seems to be a common theme to many of your error messages.Īlso, make sure that only one version of PeptideShaker is running at the same time. But if the fasta file has worked before I don't really see why it should stop working. I am using a custom database, but I'm not sure if that's causing the problem.Ĭould be the cause. Regarding MyriMatch on Linux, did you check the SearchGUI troubleshooting section? As there is a tip on MyriMatch on Linux there that could help? Before going into the issues with the loading of the data in PeptideShaker it would be nice to know why only three of the search engines worked? Could be something special with your input or setup that could be fixed? Did you get any meaningful error messages for the search engines that failed? And could you send us the SearchGUI log file as well? X!TANDEM results, as long as everything was done on the same system.
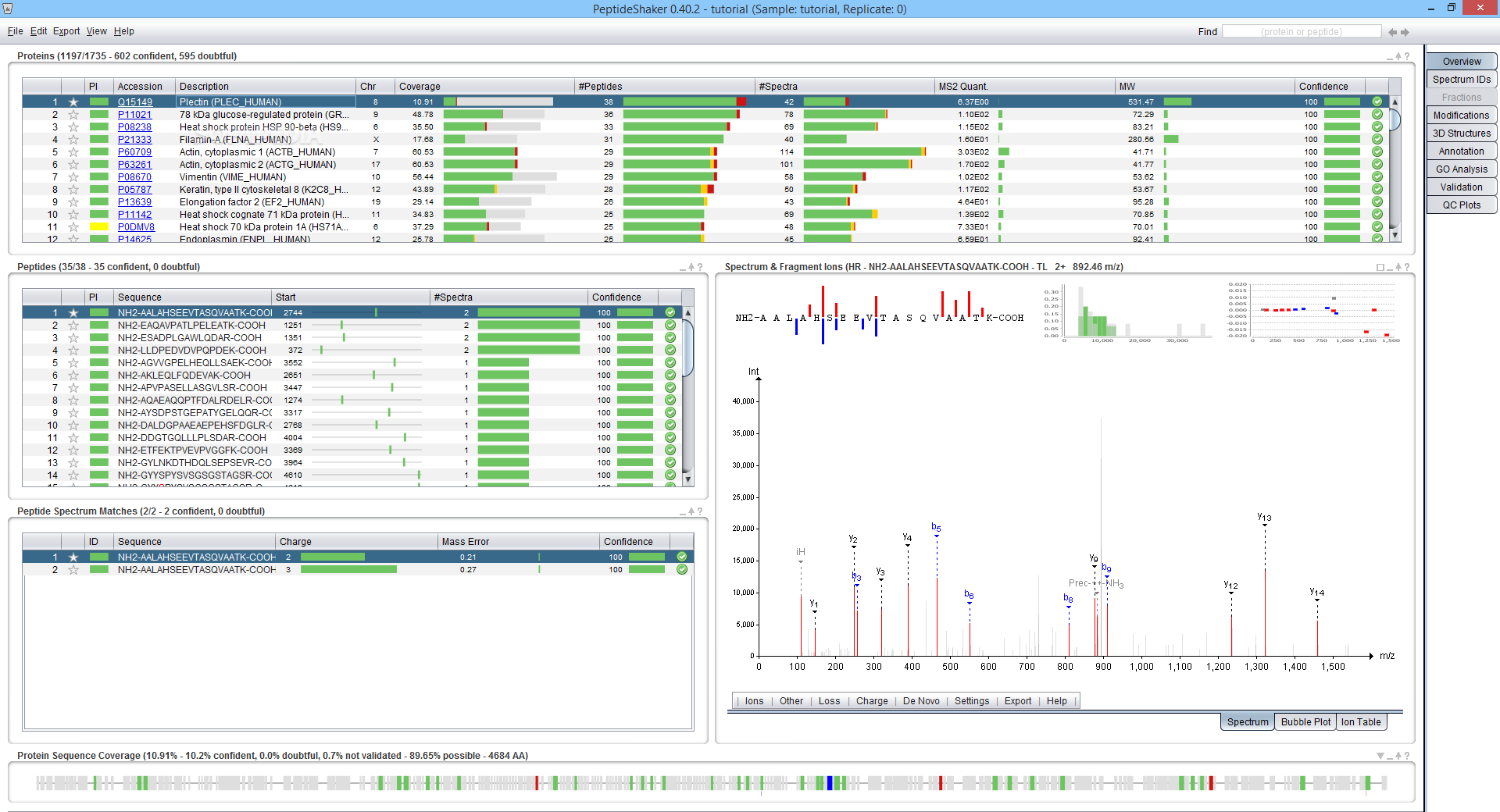
I could do the entireĪnalysis on the PC with single search engine results, and on the server with combined MS-GF+ and fasta input files successfully in the past. The MyriMatch results ("Failed to start database", "Another instance of Derby may have alreadyīooted the database", "Database could not be accessed, make sure that the file is not used by another program and that you have not run out of diskspace", something about the protein tree and something about Index out of bounds again, but this time Index and Size have the same value, "Cannot create log file at directory"). When I try the searches with each identification file individually, I get the same error messageįor MS-GF+ and X!TANDEM results (Index out of bounds), but I've seen two different messages with I also noticed that the PC and server have different versions of Java installed. I am using a custom database, but I'm not sure if par file (onlyĬhanged database and spectrum file locations). Unchanged copies of the input files, and used SearchGUI on Linux to edit the. Unfortunately, it doesn't work with the GUI or CLI of PeptideShaker on the server. I decided to try and analyse these results on our server as well (Linux it can't do the MyriMatch search in SearchGUI for some reason, only MS-GF+ and X!TANDEM, otherwise I would do everything on the server). Tried to analyse them with PeptideShaker, stating that certain database "entries were not found The computer that I used to generate these results gave an error when I
#Peptideshaker download windows 8#
Generated with SearchGUI-2.8.2 on Windows 8 - these are the only ones that work for some reason) I've been trying to analyse results from three search engines (MyriMatch, MS-GF+ and X!TANDEM,


 0 kommentar(er)
0 kommentar(er)
